Example of Parseval’s theorem¶
[1]:
import numpy as np
import numpy.testing as npt
import xarray as xr
import xrft
import numpy.fft as npft
import dask.array as dsar
import matplotlib.pyplot as plt
%matplotlib inline
First, we show that ``xrft.dft`` satisfies the Parseval’s theorem exactly for a non-windowed signal
For one-dimensional data:
Generate synthetic data
[10]:
Nx = 40
dx = np.random.rand()
da = xr.DataArray(
np.random.rand(Nx) + 1j * np.random.rand(Nx),
dims="x",
coords={"x": dx * (np.arange(-Nx // 2, -Nx // 2 + Nx)
+ np.random.randint(-Nx // 2, Nx // 2)
)
},
)
[2]:
###############
# Assert Parseval's using xrft.dft
###############
FT = xrft.dft(da, dim="x", true_phase=True, true_amplitude=True)
npt.assert_almost_equal(
(np.abs(da) ** 2).sum() * dx, (np.abs(FT) ** 2).sum() * FT["freq_x"].spacing
)
###############
# Assert Parseval's using xrft.power_spectrum with scaling='density'
###############
ps = xrft.power_spectrum(da, dim="x")
npt.assert_almost_equal(
ps.sum(),
(np.abs(da) ** 2).sum() * dx
)
For two-dimensional data:
Generate synthetic data
[11]:
Ny = 60
dx, dy = (np.random.rand(), np.random.rand())
da2 = xr.DataArray(
np.random.rand(Nx, Ny) + 1j * np.random.rand(Nx, Ny),
dims=["x", "y"],
coords={"x": dx
* (
np.arange(-Nx // 2, -Nx // 2 + Nx)
+ np.random.randint(-Nx // 2, Nx // 2)
),
"y": dy
* (
np.arange(-Ny // 2, -Ny // 2 + Ny)
+ np.random.randint(-Ny // 2, Ny // 2)
),
},
)
[3]:
###############
# Assert Parseval's using xrft.dft
###############
FT2 = xrft.dft(da2, dim=["x", "y"], true_phase=True, true_amplitude=True)
npt.assert_almost_equal(
(np.abs(FT2) ** 2).sum() * FT2["freq_x"].spacing * FT2["freq_y"].spacing,
(np.abs(da2) ** 2).sum() * dx * dy,
)
###############
# Assert Parseval's using xrft.power_spectrum with scaling='density'
###############
ps2 = xrft.power_spectrum(da2, dim=["x", "y"])
npt.assert_almost_equal(
ps2.sum(),
(np.abs(da2) ** 2).sum() * dx * dy
)
###############
# Assert Parseval's using xrft.power_spectrum with scaling='spectrum'
###############
ps2 = xrft.power_spectrum(da2, dim=["x", "y"], scaling='spectrum')
npt.assert_almost_equal(
ps2.sum() / (ps2.freq_x.spacing * ps2.freq_y.spacing),
(np.abs(da2) ** 2).sum() * dx * dy,
)
Now, we show how Parseval’s theorem is approximately satisfied for windowed data
where \(w\) is the windowing function, \(\circ\) is the convolution operator and \(\langle \cdot \rangle\) is the area sum, namely, \(\sum_x\).
Generate synthetic data: A \(300\)Hz sine wave with an RMS\(^2\) of \(200\).
[3]:
A = 20
fs = 1e4
n_segments = int(fs // 10)
fsig = 300
ii = int(fsig * n_segments // fs) # frequency index of fsig
tt = np.arange(fs) / fs
x = A * np.sin(2 * np.pi * fsig * tt)
plt.plot(tt[:100],x[:100]);
Assert Parseval’s for different windowing functions
Depending on the scaling flag, a different correction is applied to the windowed spectrum:
scaling='density': Energy correction - this corrects for the energy (integral) of the spectrum. It is typically applied to the power spectral density (including cross power spectral density) and rescales the spectrum by1.0 / (window**2).mean(). It ensures that the integral of the spectral density (approximately) matches the RMS\(^2\) of the signal (i.e. that Parseval’s theorem is satisfied).scaling='spectrum': Amplitude correction - this corrects the amplitude of peaks in the spectrum and rescales the spectrum by1.0 / window.mean()**2. It is typically applied to the power spectrum (i.e. not density) and is most useful in strongly periodic signals. It ensures, for example, that the peak in the power spectrum of a 300 Hz sine wave with RMS\(^2 = 200\) has a magnitude of \(200\).
These scalings replicate the default behaviour of scipy spectral functions like scipy.signal.periodogram and scipy.signal.welch (cf. Section 11.5.2. of Bendat & Piersol,
2011; Section 10.3 of Brandt,
2011).
[4]:
RMS = np.sqrt(np.mean((x**2)))
windows = np.array([["hann","bartlett"],["tukey","flattop"]])
Check the energy correction for ``scaling=’density’``: With window_correction=True, the spectrum integrates to RMS\(^2\).
[5]:
fig, axes = plt.subplots(figsize=(13,10), nrows=2, ncols=2)
fig.set_tight_layout(True)
for window_type in windows.ravel():
x_da = xr.DataArray(x, coords=[tt], dims=["x"]).chunk({"x": n_segments})
ps = xrft.power_spectrum(
x_da,
dim="x",
window=window_type,
chunks_to_segments=True,
window_correction=True,
).mean("x_segment")
ps.plot(ax=axes[np.where(windows==window_type)[0][0],np.where(windows==window_type)[1][0]])
axes[np.where(windows==window_type)[0][0],np.where(windows==window_type)[1][0]].set_title(window_type, fontsize=15)
npt.assert_allclose(
np.trapz(ps.values, ps.freq_x.values),
RMS**2,
rtol=1e-3
)
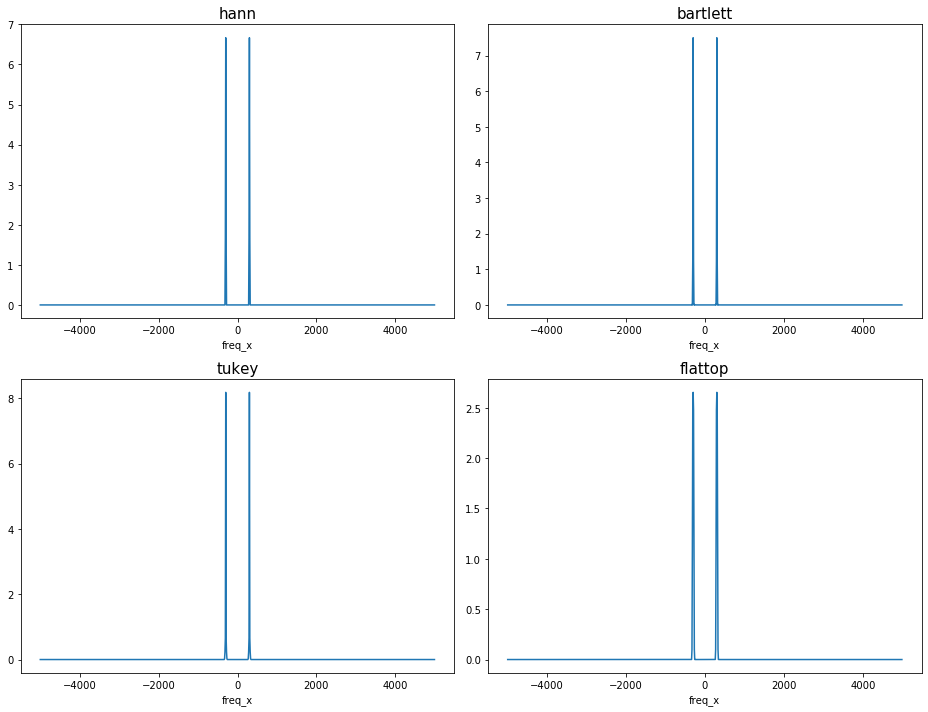
The maximum amplitude differs amongst cases with different windows due to the difference in noise floor.
Check the amplitude correction for ``scaling=’spectrum’``: With window_correction=True, the peak of the two-sided spectrum has a magnitude of RMS\(^2/2\).
[6]:
fig, axes = plt.subplots(figsize=(13,10), nrows=2, ncols=2)
fig.set_tight_layout(True)
for window_type in windows.ravel():
x_da = xr.DataArray(x, coords=[tt], dims=["x"]).chunk({"x": n_segments})
ps = xrft.power_spectrum(
x_da,
dim="x",
window=window_type,
chunks_to_segments=True,
scaling="spectrum",
window_correction=True,
).mean("x_segment")
ps.plot(ax=axes[np.where(windows==window_type)[0][0],np.where(windows==window_type)[1][0]])
axes[np.where(windows==window_type)[0][0],np.where(windows==window_type)[1][0]].set_title(window_type, fontsize=15)
# The factor of 0.5 is there because we're checking the two-sided spectrum
npt.assert_allclose(ps.sel(freq_x=fsig), 0.5 * RMS**2)
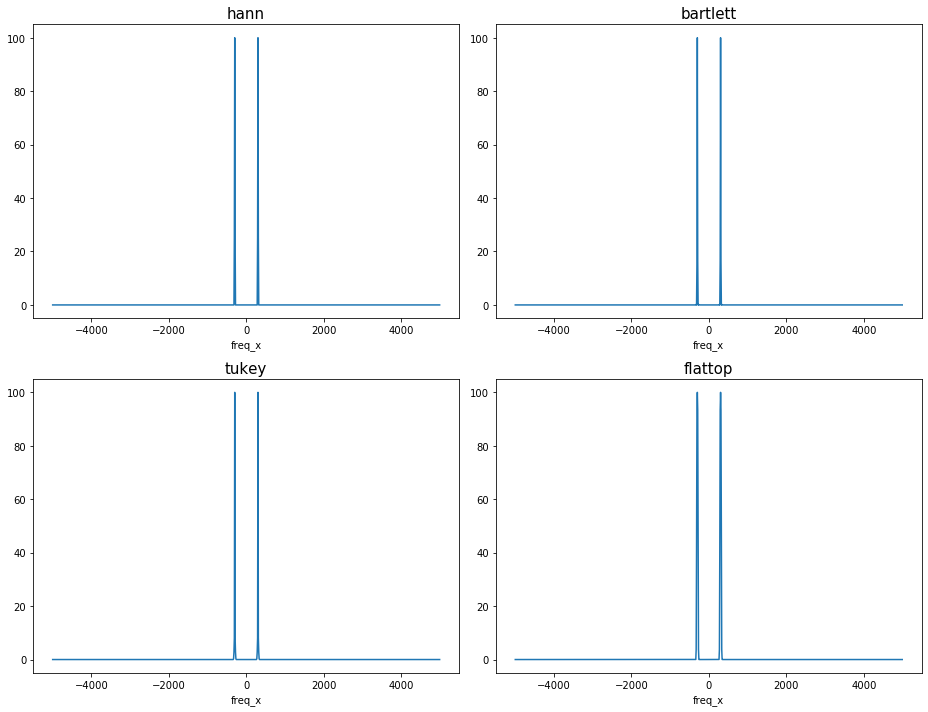
The maximum amplitudes are now all the same amongst different windows applied with the amplitude correction.
[ ]: